The MFDB system
Generally when databases are mentioned, one envisages a central server to connect to, acting as a hub for information storage. Information is stored and then searched, combining together as appropriate. Thanks to advances in computing power, we do not need to install databases onto specialist servers. We can install a database onto our own laptop, and still take advantage of the data transformation features they offer.
The MareFrame Database is not a central database. Instead it is a R package, or toolbox, to help you manage a database on your own computer. It provides:
- Automatic set-up and configuration of a PostgreSQL database
- Functions to ingest data automatically from files or other database APIs
- Functions to transform and aggregate the data
- Functions to create input files for ecosystem modelling tools, e.g. GADGET and RPath
The advantages of this approach are:
- No specialist database knowledge required; MFDB sets up the database tables for you, and creates all queries.
- Unlike a spreadsheet or flat files, the database always checks that data matches the required form and will error otherwise. It is also impossible to e.g. copy and paste a bunch of rows mistakenly.
- Built-in knowledge of ecosystem modelling tools, data file output will always be valid.
- Model creation can be automated using R, speeding up model development time, making updating the model with new data easy, and ensuring repeatablility.
Overview
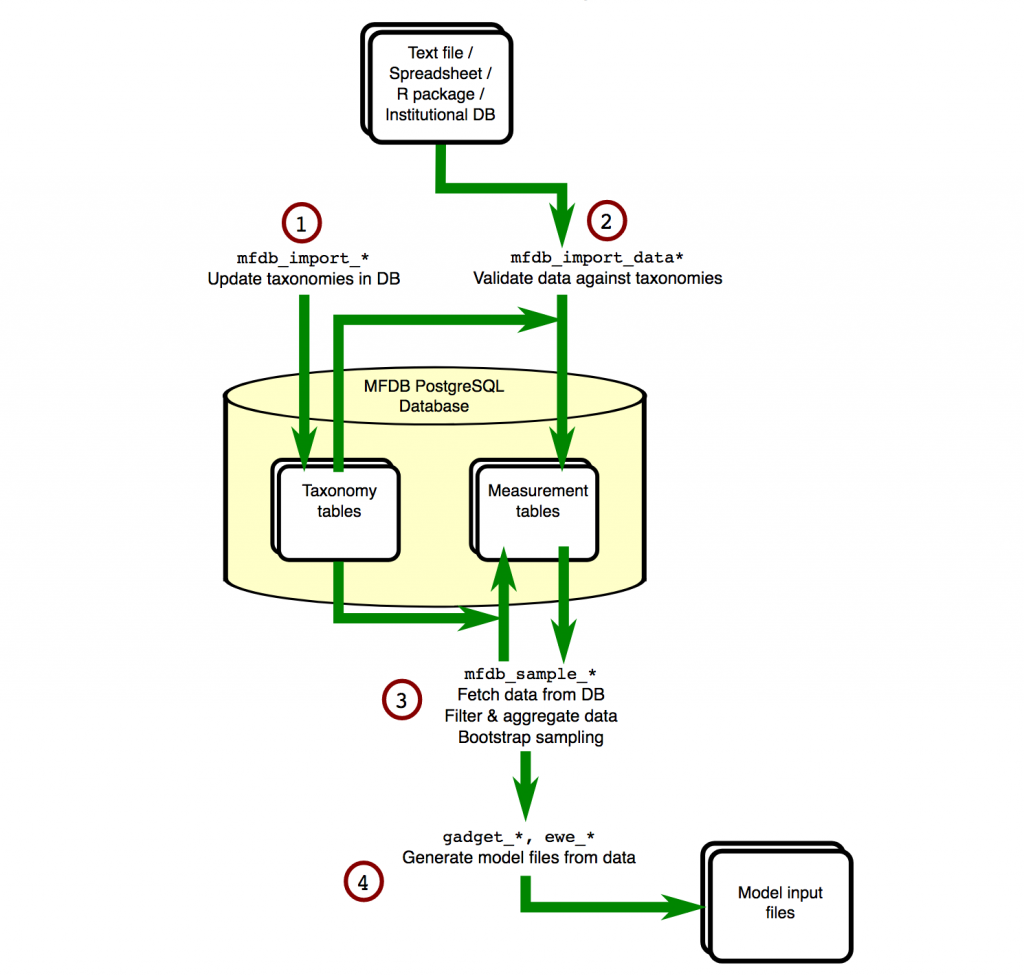
The flow of information around an MFDB database is shown in the diagram below:
The first step is defining taxonomies. For many aspects of the data, e.g. area, species, vessel, tows, any possible values have to be defined in advance. This means data can be checked on entry, and if it doesn’t match then errors can be reported.
Next we import the actual data (2). The database is designed for marine ecosystem modelling, and so the concepts it can store are related. For example…
- Data from survey fleets, commercial landings logbooks, etc.
- Stomach content surveys
- Surface temperature data and other survey indices, e.g. acoustic data
Because we are using the R language, we can either use R’s built-in commands to download & read files, or other R packages to interface to APIs that make data available. We do not need particular file formats, as long as R can read it.
Now, queries can be made (3). You do not need to write the queries yourself, MFDB provides functions that generate reports, all of which can…
- Filter data, for example by species and areas.
- Aggregate data, for example into length groups
- Perform bootstrap sampling of data, for example by area.
Finally, the output of these queries can be used to automatically create model input files (4). In the case of GADGET, all associated files, e.g. area and length aggregation files, will be generated at the same time, using the same information you used to query MFDB.
Getting more information
The package is available and developed at Github – http://github.com/mareframe/mfdb/. Here you can dowload the source, view documentation and report any issues you find.
The package is fully documented with standard R package documentation, and has a suite of verified-working demos to demonstrate aspects of usage. The HTML version of the documentation is available online.
All functions have R help pages. Once installed you can read an overview of using MFDB by using the package?mfdbcommand. All other MFDB commands have documentation, you can see a list by using the ??mfdb command.
There are example scripts that can import from ICES and MRI databases included in the MFDB package. These can be either run using the demo(package = 'mfdb') command, or you can look at the demo code on GitHub.